9+ Calculate_Qc_Metrics
For planning purposes the Patient Risk Sigmas used for the calculation of run sizes are 528. The calculator results are shown below.

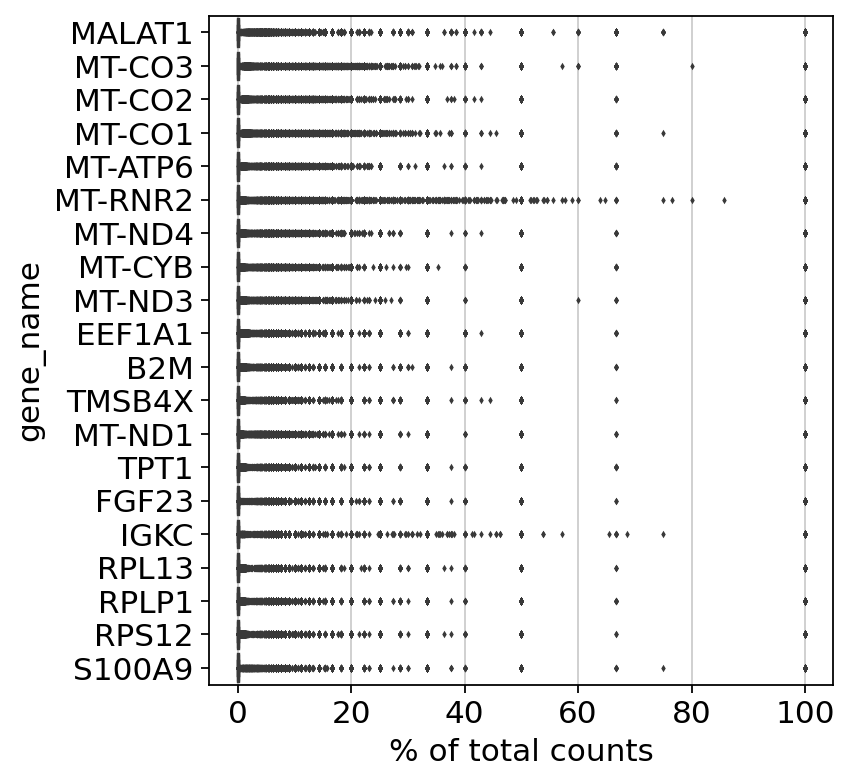
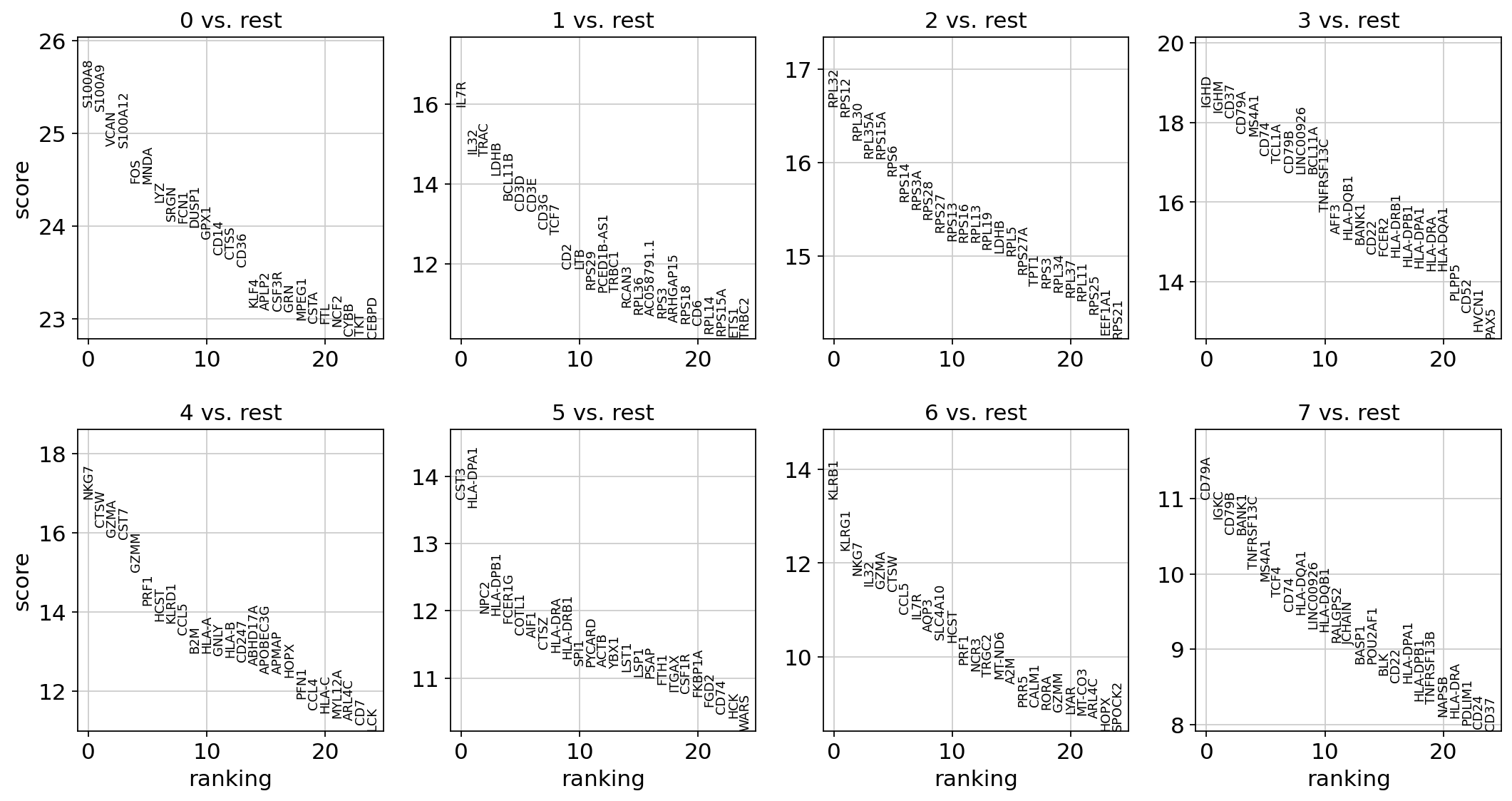
Analysis And Visualization Of Spatial Transcriptomics Data Scanpy Documentation
There are 3 main areas where QC can be applied to NGS.

. The calculated sigmas are 528 909 and 939 resp. Automatically run every analysis. Calculate useful quality control metrics to help with pre-processing of data and identification of potentially problematic features and cells.
Hi Thanks for making Scanpy. We calculate the QC metrics described above with a combination of methods from the scater McCarthy et al. By voting up you can indicate which examples are most useful and appropriate.
2017 package for metrics that are also used for. The Figure gives a graphical example of a test method with 1 bias 25 coefficient. Import gene annotation anno_to_saf.
Here are the examples of the r api scater-calculateQCMetrics taken from open source projects. The output Defaults to 8file of each analysis will start with this prefix name. Datasc_example_counts datasc_example_cell_info pd.
I think this might be best raised an issue over on the numba repositorygiovp. Contribute to LuyiTianscPipe development by creating an account on GitHub. Calculate_qc_metrics rna qc_vars mito inplace True Yes would be great to the see the QC plots here too.
Genestsv matrixmtx and barcodestsv. Im not sure how much I can help with this since I dont have a windows machine to try this on. It is a zip file containing the market matrix files that I showed you in class.
Quality Control QC metrics can be viewed in QC reports and in final reports. This is required if no individual metrics are specified. The metrics are based on data that are included in an analysis.
Where CV is the analytical coefficient of variation of the test method. The following QC metrics are computed. Calcuate QC metrics from gene count matrix.
31 rows Quality Control QC metrics. Sigma TEa - Bias CV. It works wonderfully and is in general easy to use.
A pipeline for single cell RNA-seq data analysis. You will need to unzip it and then change the path argument below so that it. On the starting nucleic acids After Library preparation Post-Sequencing Ultimately the best QC of your NGS experiments is.
Convert annotation from GenomicRanges to Simple Annotation. We can look check out the qc metrics for our data. However I am confused by the exact meaning of QC metrics as calculated by.
94 Calculate QC metrics.
Python Kallisto Bustools
Python Kallisto Bustools
Python Kallisto Bustools

Analysis And Visualization Of Spatial Transcriptomics Data Scanpy Documentation

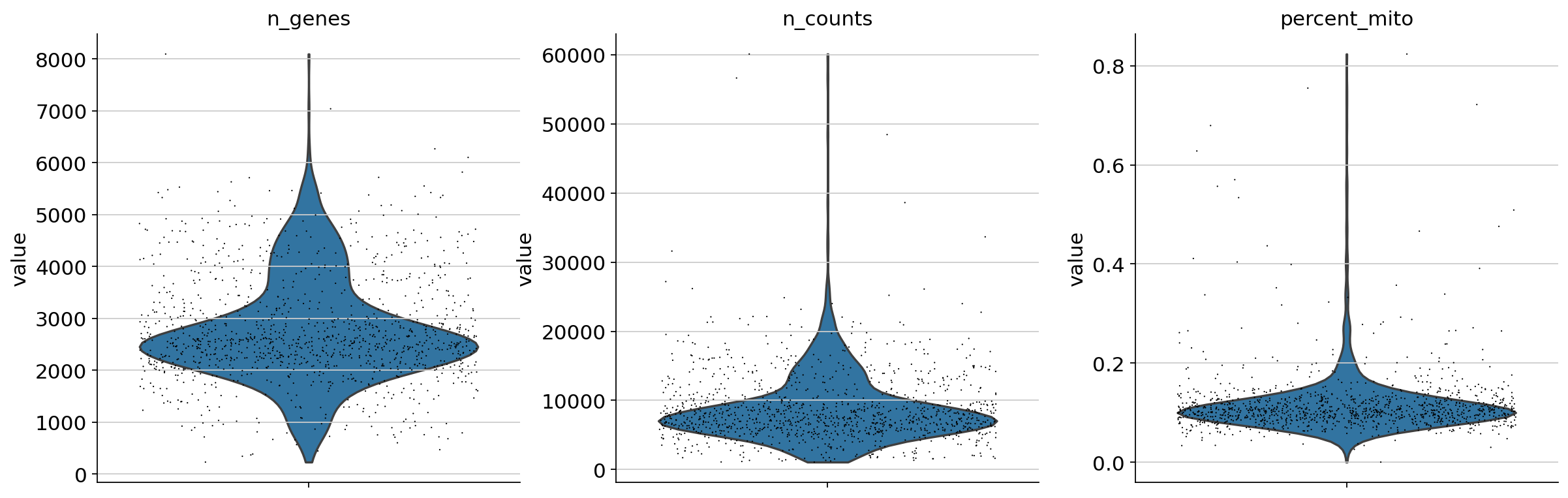
Scanpy 01 Qc
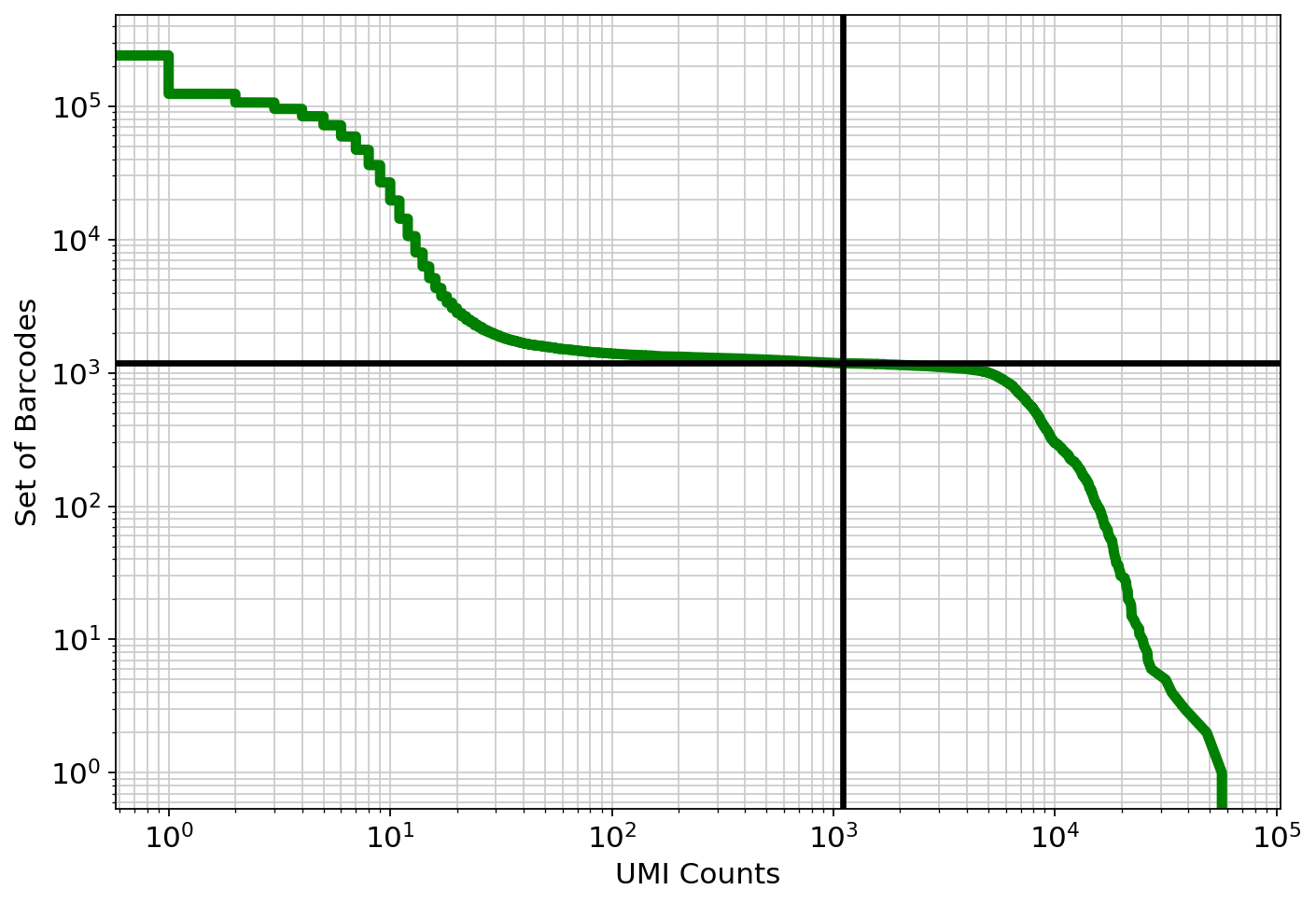
Python Kallisto Bustools

Scanpy 01 Qc
Python Kallisto Bustools
Python Kallisto Bustools
Python Kallisto Bustools

Scanpy 01 Qc

Scanpy 01 Qc

Latest Help Topics Scanpy

Analysis And Visualization Of Spatial Transcriptomics Data Scanpy Documentation
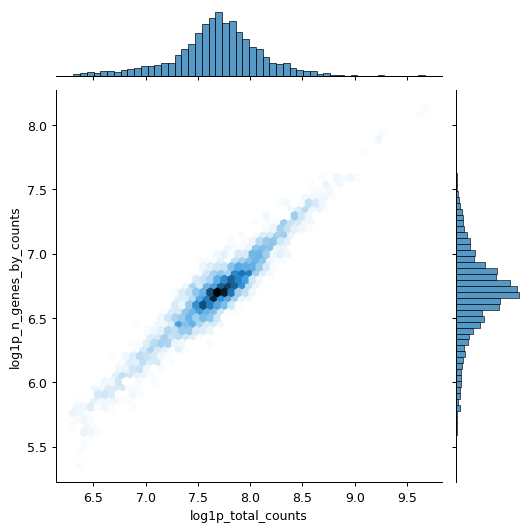
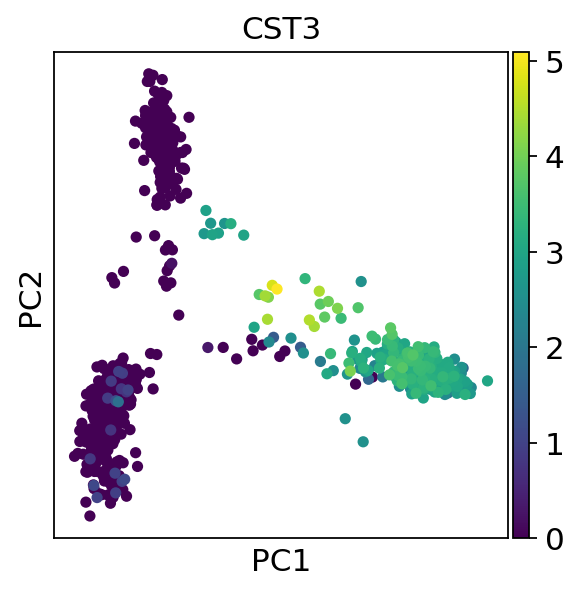
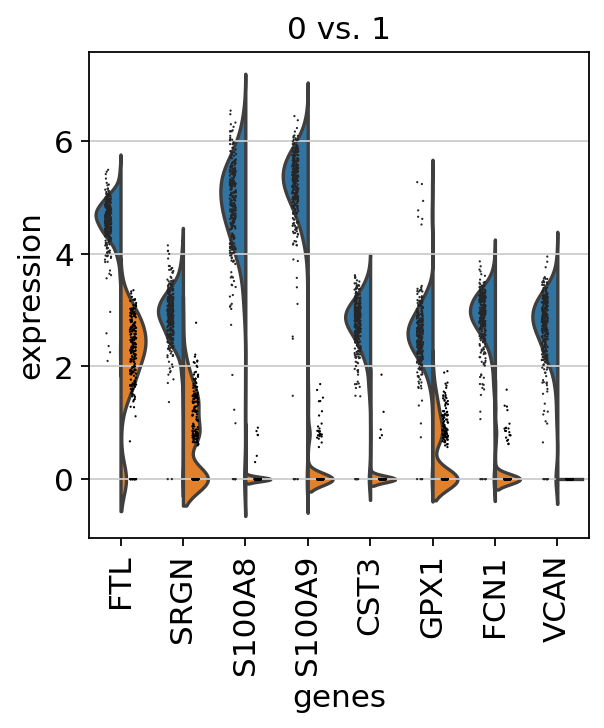
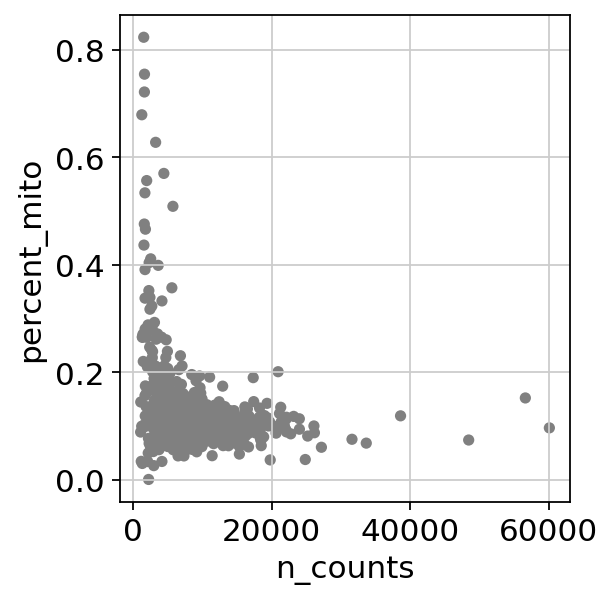
Scanpy Pp Calculate Qc Metrics Scanpy 1 9 1 Documentation
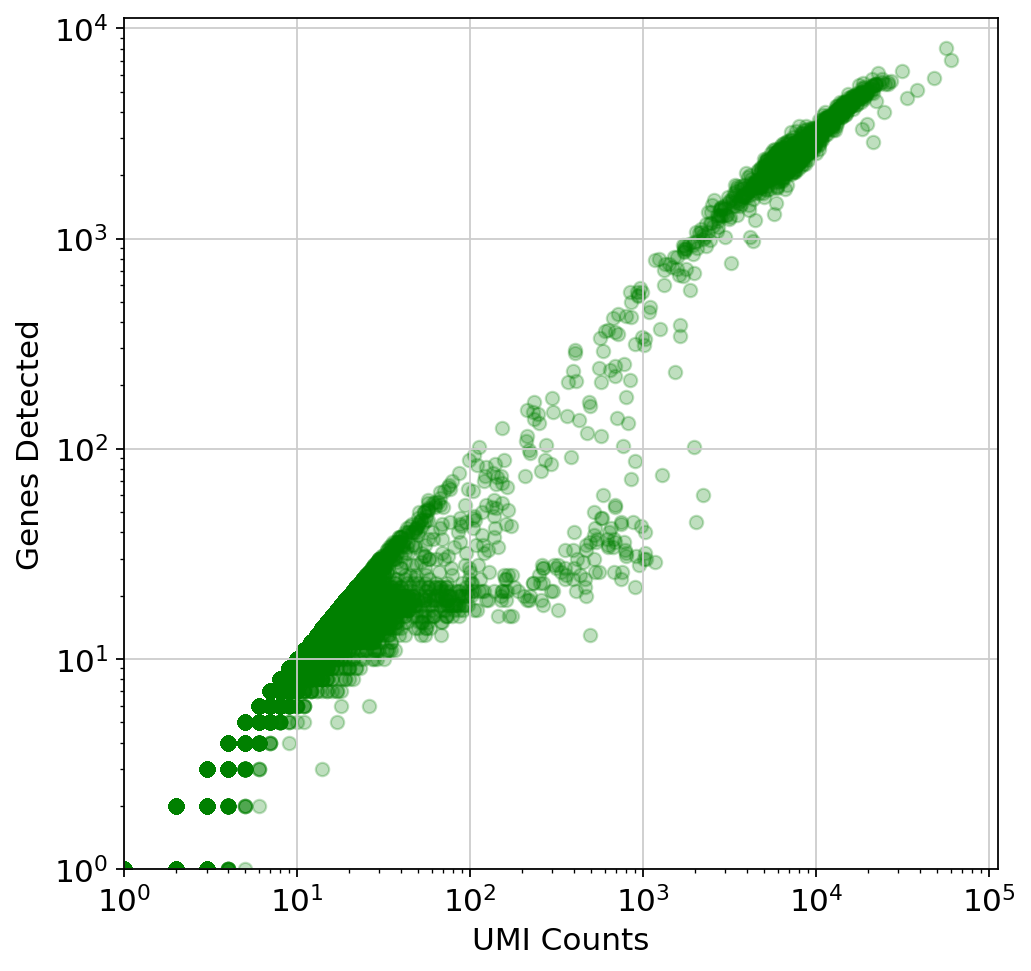
Python Kallisto Bustools

Analysis And Visualization Of Spatial Transcriptomics Data Scanpy Documentation